Big Self-Supervised Models Advance Medical Image Classification
Paper Summary
Paper
TL;DR
-
Objective is to learn from limited labeled data in a medical image analysis setting, that are robust to distribution shifts. They use SimCLR not SimCLRv2.
-
They study the effectiveness of self-supervised learning as a pre-training strategy in medical image classification. They perform comparison between supervised pre-training and self-supervised pre-training approaches.
- Tasks
- Dermatology condition classification
- Multi-label chest x-ray classification
- Intuitions and Key Ideas:
-
Self-supervised learning on ImageNet, followed by additional self-supervised learning on unlabeled domain-specific medical images significantly improves the accuracy of medical image classifiers.
-
Big self-supervised models are robust to distribution shift and can learn efficiently with a small number of labeled medical images.
-
MICLe - uses multiple images of the underlying pathology per patient case to construct more informative positive pairs for self-supervised learning.
-
We observe that self-supervised pretraining outperforms supervised pretraining, even when the full ImageNet dataset (14M images and 21.8K classes) is used for supervised pretraining. We attribute this finding to the domain shift and discrepancy between the nature of recognition tasks in ImageNet and medical image classification. Self-supervised approaches bridge this domain gap by leveraging in-domain medical data for pretraining.
-
Multi-Instance Contrastive Learning (MICLe)
-
strategy that helps adapt contrastive learning to multiple images of the underlying pathology per patient case. Such multi-instance data is often available in medical imaging datasets – e.g., frontal and lateral views of mammograms, retinal fundus images from each eye, etc.
-
Given multiple images of a given patient case, they construct a positive pair for self-supervised contrastive learning by drawing two crops from two distinct images of the same patient case. These images are typically from different viewing angles and show different body parts with the same underlying pathology. This enables self-supervised learning algorithms to learn representations that are robust to changes of viewpoint, imaging conditions, and other factors in a direct way.
-
MICLe does not require class label information.
-
-
Key Ideas
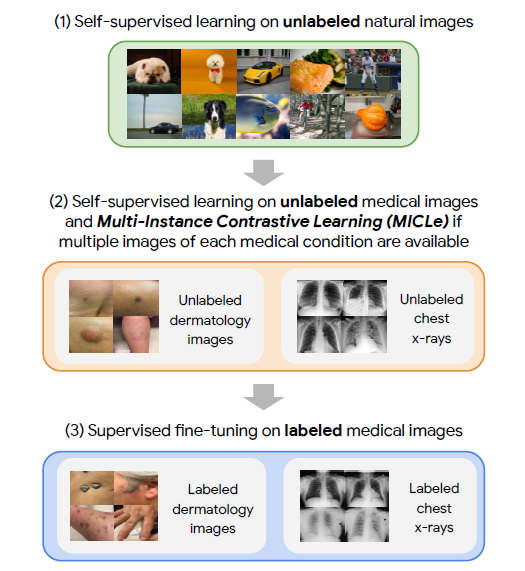
- Their approach comprises of three steps:
-
Self-supervised pretraining on unlabeled ImageNet using SimCLR
-
Additional self-supervised pretraining using unlabeled medical images. If multiple images of each medical condition are available, a novel Multi-Instance Contrastive Learning (MICLe) is used to construct more informative positive pairs based on different images.
-
Supervised fine-tuning on labeled medical images. Note that unlike step (1), steps (2) and (3) are task and dataset specific.
-
-
First, they perform self-supervised pretraining on unlabeled images using contrastive learning to learn visual representations.
-
For contrastive learning they use a combination of unlabeled ImageNet dataset and task specific medical images. Then, if multiple images of each medical condition are available the Multi-Instance Contrastive Learning (MICLe) is used for additional self-supervised pretraining.
- Finally, we perform supervised fine-tuning on labeled medical images.
- MICLe
- It is common for medical images to have multiple images per condition typically from different viewpoints, lighting conditions, providing complementary information.
- To get 2N representations in MICLe, they randomly sample a mini-batch of N bags of instances and define the contrastive prediction task on positive pairs retrieved from the bag of images instead of augmented views of the same image.
- Each bag
X = {x₁, x₂, ..., x_M}has images from the same patient (i.e., same pathology) captured from different views, whereMcan vary across bags.
When there are two or more instances in a bag (M = |X| ≥ 2), positive pairs are constructed by drawing two crops from two randomly selected images in the bag.
In this case, the objective still takes the form of SimCLR, but the images contributing to each positive pair are distinct.
-
Following SimCLR, two fully connected layers are used to map the output of ResNets to a 128-dimensional embedding, which is used for contrastive learning.
-
SimCLR pre-training is performed on unlabeled dermatology/chest X-ray samples both with and without initialization from pre-trained ImageNet self-supervised weights.
- Augmentations
-
Dermatology - same augmentation as SimCLR.
- Chest Xray
-
Unlike the original set of proposed augmentation in SimCLR, they do not use the Gaussian blur, because they deduce that it makes it impossible to distinguish local texture variations and other areas of interest thereby changing the underlying disease interpretation the X-ray image.
-
Augmentations that lead to the best performance on the validation set for this task are random cropping, random color jittering (strength = 0.5), rotation (upto 45 degrees) and horizontal flipping.
-
- Finetuning
- For data augmentation during fine-tuning, they used random color augmentation, crops with resize, blurring, rotation, and flips for the images in both tasks.
-
Evaluation
-
The primary metrics for the dermatology task are top-1 accuracy and Area Under the Curve (AUC)
-
For the chest X-ray task, given the multi-label setup, they report mean AUC averaged between the predictions for the five target pathologies.
- They investigate three possible scenarios for self-supervised pretraining in the medical context:
- (1) using ImageNet dataset only
- (2) using the task specific unlabeled medical dataset (i.e. Derm and CheXpert)
- (3) initializing the pretraining from ImageNet self-supervised model but using task specific unlabeled dataset for pretraining, indicated as ImageNet→CheXpert and ImageNet→CheXpert
- Testing on OOD data
- They use the model post pretraining and end-to-end fine-tuning (i.e. CheXpert and Derm) to make predictions on an additional shifted dataset without any further fine-tuning (zero-shot transfer learning). Dexternal-Derm and DNIH are used as target shifted datasets.
Observations
-
They note that when only using ImageNet for self-supervised pretraining, the model performs worse in this setting compared to using in-domain data for pretraining.
-
They show that self-supervised models are robust and generalize better than baselines, when subjected to shifted test sets, without fine-tuning.
- Testing label efficiency of Self-supervised learning
- They fine-tune models on different fractions of labeled training data and also conduct baseline finetuning experiments with supervised ImageNet pretrained models. They use label fractions ranging from 10% to 90% for both Derm and CheXpert training datasets.
- MICLe yields proportionally larger gains when fine-tuning with fewer labeled examples. MICLe is able to match baselines using only 20% of the training data for ResNet-50 (4×) and 30% of the training data for ResNet-152 (2×).
Enjoy Reading This Article?
Here are some more articles you might like to read next: